Screencast
Explore
The Explore section of fusionDB enables you to search for organisms annotated and stored in fusionDB. The search field (1) let's you combine search terms like organism name, habitat preferences and oxygen preferences separated by space. The search logic is fuzzy i.e. it looks for occurrences of strings within the table (not case sensitive). As a result a simple search for "Synechococcus" will also retrieve "Thermosynechococcus". To circumvent this search terms can be surounded by quotation marks including a leading or tailing whitespace. If we would like to retrieve only Synechococcus with habitat "fresh water" in the filtered organisms we would have to search for '" Synechococcus" fresh water'. Note that since the Search field only contains "Synech fresh" Synechocystis and Thermosynechococcus are as well returned.
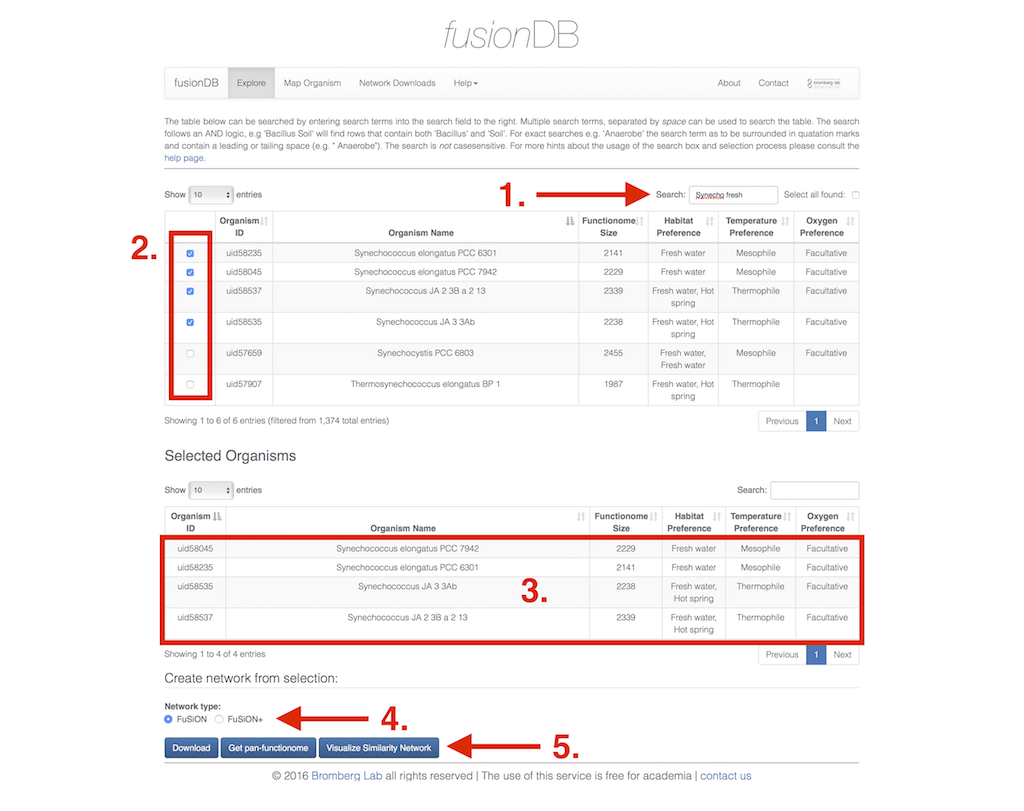
When you have found the organisms of your interest you can use the checkboxes (2) in the first column of the table to select the microbes that you want to investigate further. Note that removing the search term from the search field maintains the selection just made. Hence, you can repeat using the search field as often as you like without loosing your current selection. To select all organisms of your current search you can click the checkbox next to the search field. Likewise to deselect the whole selection. Upon selection the organisms will be displayed in a second selected organism table (3) for easier overview. Deselecting organisms will remove them again.
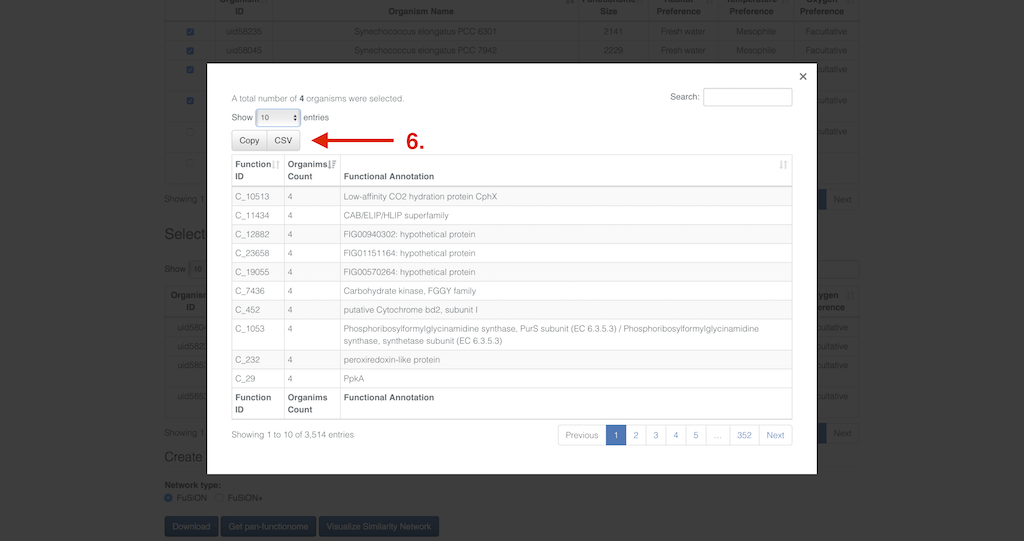
As soon as you have finished the selection you have multiple options how to proceed further. First you can select the type to network created from the selected organisms you want to download - fusion or fusion+ (4). This selection also applies for the possible in browser Visualization (5) of the network. The download button will create a popup in which you can decide upon the file format (abc & gexf). Additionally you can get the pan functional repertoire of the selected organisms. Clicking the button will spawn a popup as well. This popup displays a table containing the function id, the number of organisms that share the function, and the functional annotation. The list can also be search and downloaded (6) as comma separated value file or copied into the clipboard.
Map Organims
The second functionality that fusionDB offers is Mapping a organism's proteome to the reference functional reptertoire extracted from 1374 organisms. There are two ways of providing the proteome (in FASTA-format): Pasting the sequences into the text field (1) prominently placed in the interface or uploading (2) the multi fasta-file. Additionally, if an optional email (3) address is provided, you will be notified by email when the job is finished including a link that will direct you directly to the results page.

As soon as the mapping job is submitted to the cluster back-end you will be forwarded to a status page that displays the progress (4) of the job and additional information. If no email address was provided this page should be booked marked since you can be redirected (5) to the results page from here as soon as the job is finished.
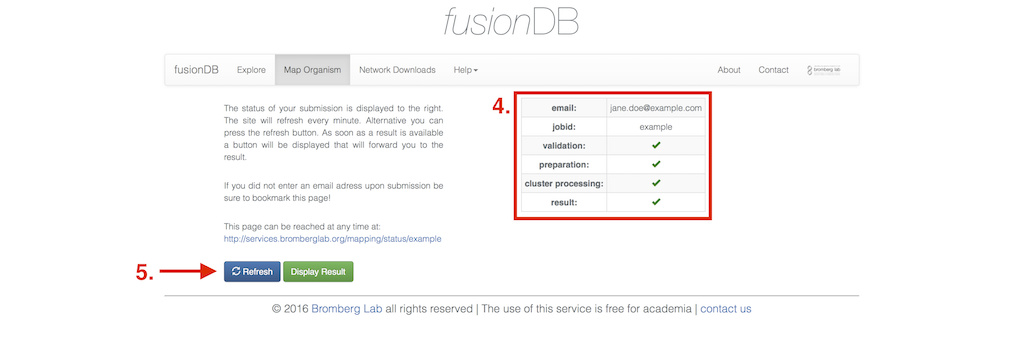
The results page contains general information (6) about the mapped proteome, i.e. how many proteins where mapped to how many functions, and how many proteins were assigned as singletons. The page contains a Functions table listing all the functions the proteome was mapped to, including the functional annotation, and a count how many proteins where mapped to that specific function (8). Clicking on the info button in this column will spawn a pop-up that lists the mapped proteins by their identifier given in the uploaded multi-fasta. The pop-up also contains a possibility to download a multi-fasta file containing the protein sequences of those mapped proteins.
The function table is search-able (7) with the same syntax as the organism table in Explore, i.e. fuzzy search, AND logic separating single search terms with space. On top this we also offer the possibility to download the full table of the functional mapping as comma separated value file or copy the contents to the clipboard (9) - this functionality is also provided for the organism similarity table.
Further down the Organism Similarities table can be found. This table essentially displays the same functionality as the organisms table in Explore with the exception of additional information displayed (10) and the fact that the pan functional repertoire, download and visualization of the fusion(+) network generated from the selected organisms also contains the query organism.

Visualization
Initiating the visualization of selected type of similarity network will redirect after a moment to the Visualization (1). The displayed network is a prerendered layout of the selected organisms according to their location in the full fusion network. The standard coloring upon arrival is a coloring by phylum. The current coloring scheme is listed in the legend (2). In this example the nodes of the network have been recolored (3) according the organism's temperature preference. Additional options are all taxonomic levels from phylum down to the species level as well as four more types of metadata (habitat, oxygen, salinity and ph value preference). The displayed network of selected organisms can be download (4) as either rendered images (png/pdf) or network files (gexf/gml) that can be used for your own visualization purposes. Finally the network can be investigated interactively (5).
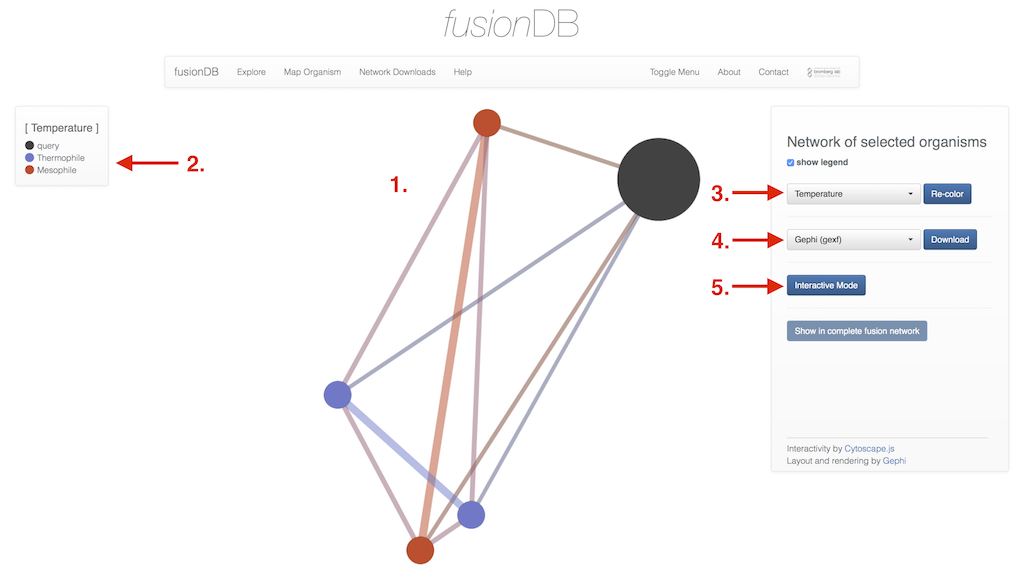
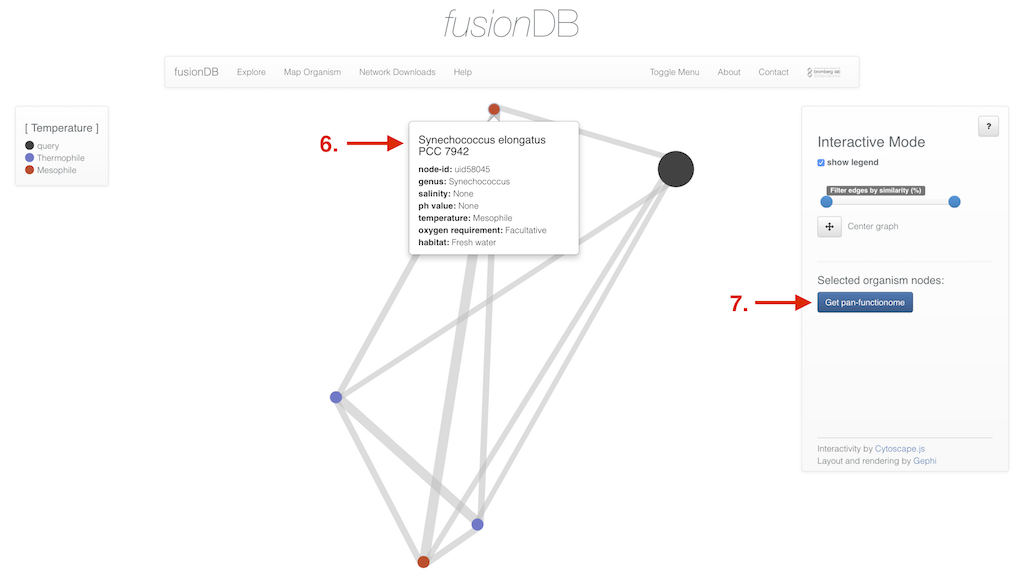
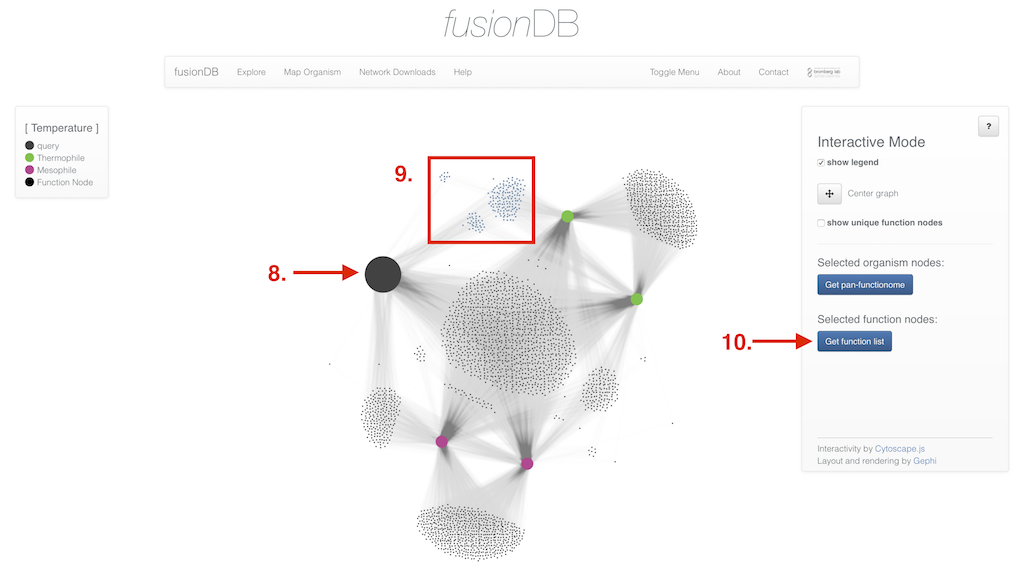
In the interactive view of the network you have multiple additional options depending on which network type was created. In the interactive display of a fusion network organisms nodes reveal further information upon click (6). Edges can be clicked as well and enable you (besides displaying the organism similarity) to retrieve the pan functional repertoire of the two organisms connected. Additionally multiple organism nodes can be selected holding down a modifer key to retrieve their pan functional repertoire (7). If the selected network type was fusion+ the interface enables you to not only select organism nodes (8) but also functional nodes (9) and retrieve their functional annotation (10).