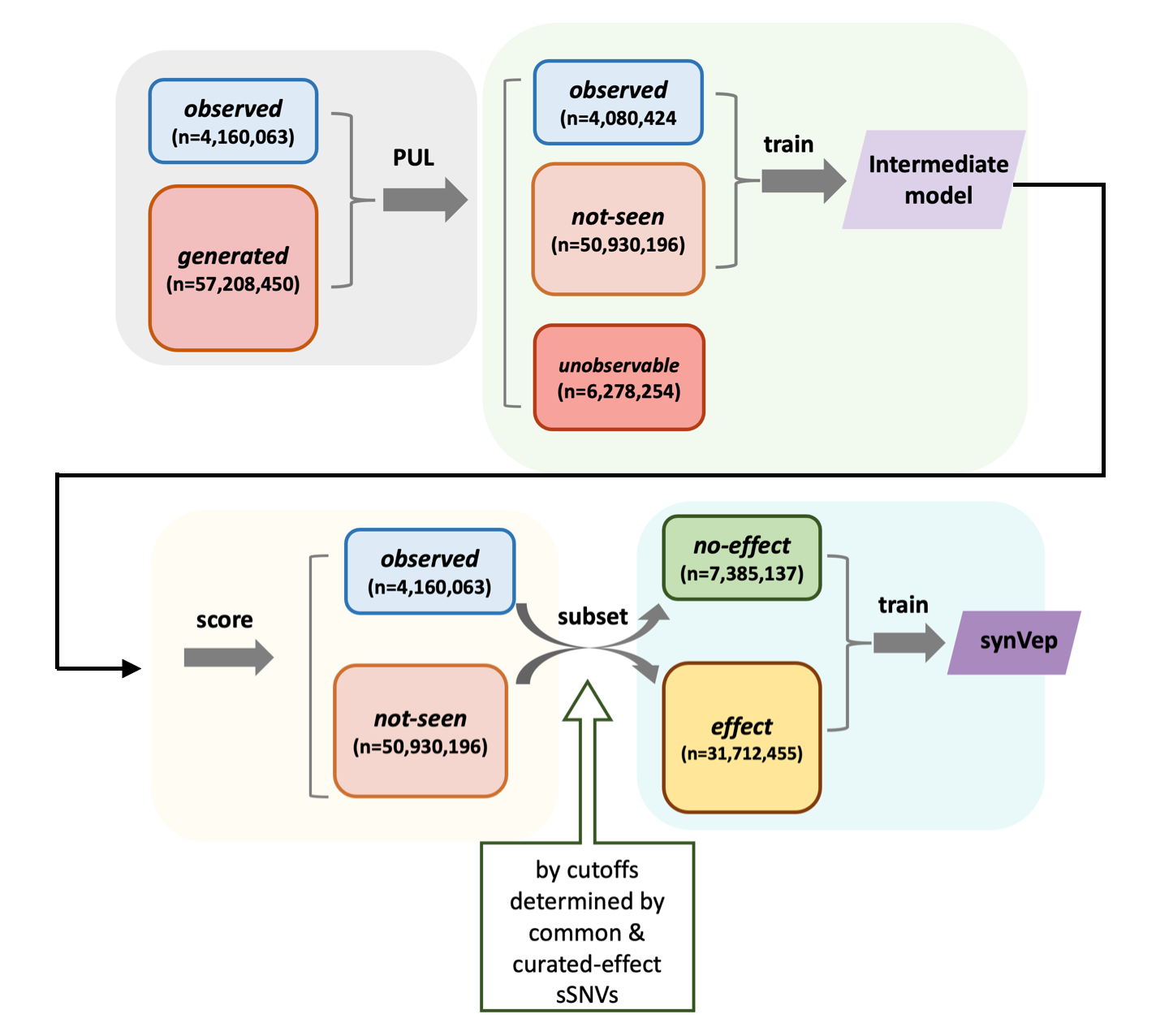
synVep is a machine learning-based predictor for evaluating the effects of human synonymous variants. It comprises a series of three models using observed (gnomAD) and generated (all possible, but not seen in gnomAD) variants: a positive-unlabeled learning (PUL) model to identify “unobservable” variants from the generated set (the observable generated are then referred to as not-seen); an intermediate model to differentiate the observed and not-seen; and a final model to classify putative effect and no-effect variants. The synVep prediction is the probability of a human synonymous variant having an effect; it’s correlation with the magnitude of effect has not yet been evaluated. For more information, please refer to the synVep paper.
If you find synVep useful in your research, please cite:
Zeng, Zishuo, Ariel A. Aptekmann, and Yana Bromberg. "Decoding the effects of synonymous variants." bioRxiv (2021).
(link)