About
Microbial functional diversification is driven by environmental factors, i.e. microorganisms inhabiting the same environmental niche tend to be more functionally similar than those from different environments. In some cases, even closely phylogenetically related microbes differ more across environments than across taxa. While microbial similarities are often reported in terms of taxonomic relationships, no existing databases directly links microbial functions to the environment.
fusionDB is a database using functional data to represent 1,374 taxonomically distinct bacteria annotated with available metadata: habitat/niche, preferred temperature, and oxygen use. Each microbe is encoded as a set of functions represented by its proteome and individual microbes are connected via common functions. Users can search fusionDB via combinations of organism names and metadata (Explore). Moreover, the web interface allows mapping new microbial genomes to the functional spectrum of reference bacteria (Map Organism), rendering interactive similarity networks that highlight shared functionality.
fusionDB is a service that is provided free for academic use. If you use the service please cite:- fusionDB: Zhu C, Mahlich Y, Miller M, Bromberg Y (2018) fusionDB: assessing microbial diversity and environmental preferences via functional similarity networks. Nucleic Acids Research, Volume 46, Issue D1, 4 January 2018, Pages D535–D541, doi: 10.1093/nar/gkx1060
- Fusion: Zhu C, Delmont TO, Vogel TM, Bromberg Y (2015) Functional Basis of Microorganism Classification. PLOS Computational Biology 11(8): e1004472. doi: 10.1371/journal.pcbi.1004472
Changelog
January 2017: Redesign of fusionDB interface
We are prod to announce the redesign of fusionDBFebraury 2016: Major changes to strictness of HSSP cut-off
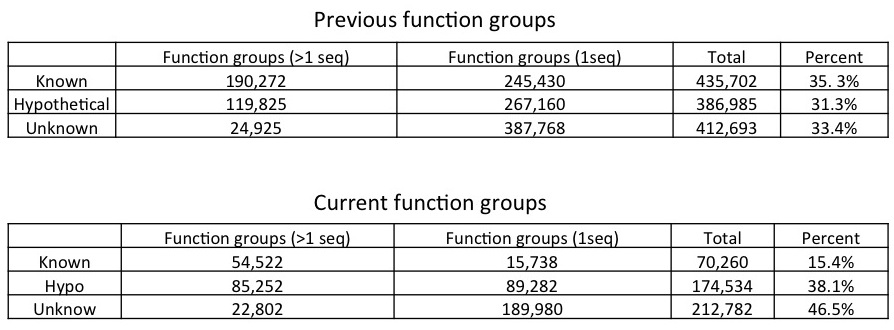
We now use a more loose cutoff to define our function groups. The comparison is shown in the picture below. For a given pairwise protein alignment, we plot it in the two dimensional space with alignment length as X axis and percent identity as Y axis. Alignments above curve are considered as sharing the same function (1). With the current curve (green) we include more same-function alignments than previous (black curve).

The function groups we defined from our bacterial genomes are thus affected - we now have fewer function groups with more protein members. The actual change is shown in th tables below.
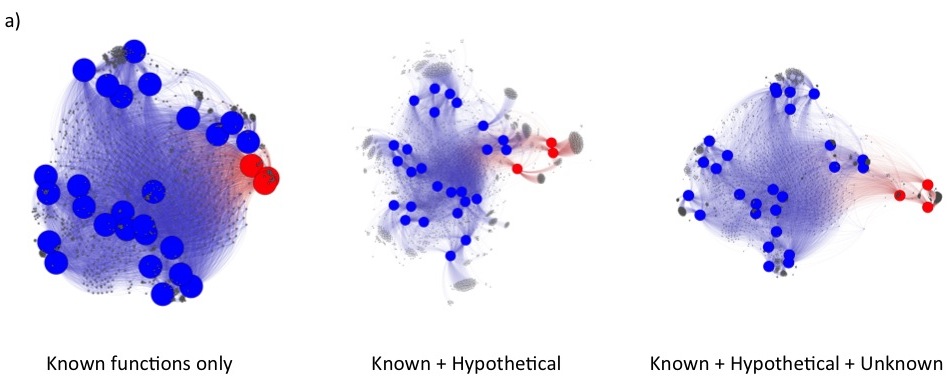
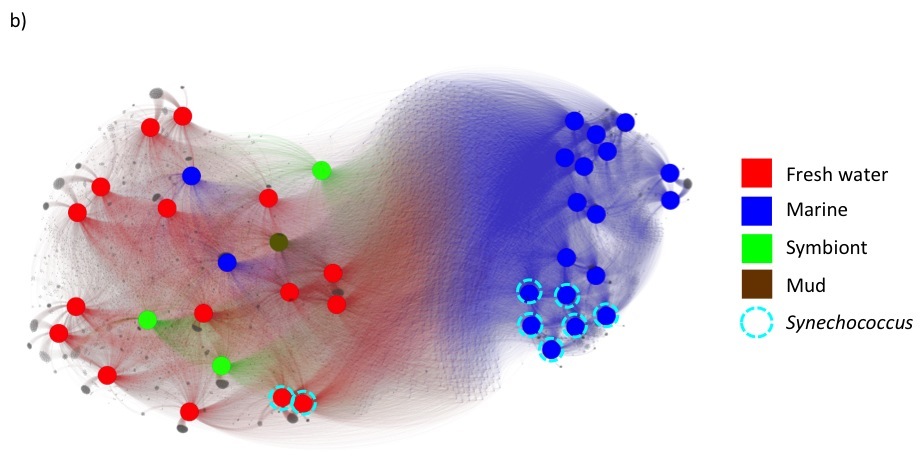
Under these changes, fusion+ generated from the current fusionDB keeps its power in revealing environment impact on bacterial functions as shown below. In a), unique blood Mycoplasma organisms are indicated by red nodes, with the rest of Mycoplasma colored in blue. Note the resolution increases as hypothetical and unknown functions added; In b), The Cyanobacteria form one mostly fresh water cluster and one marine cluster. The members of Synechococcus exist in both clusters. See C. Zhu et. al. (2) for more details of interpretation of the figures.
- Rost B (2002) Enzyme function less conserved than anticipated. J Mol Biol 318: 595-608.
- Zhu C, Delmont TO, Vogel TM, Bromberg, Y. (2015) Functional Basis of Microorganism Classification. PLoS Comput Biol, 11, e1004472.